Open access preprint (which I announced already) at bioRxiv Patterns of genetic differentiation and the footprints of historical migrations in the Iberian Peninsula, by Bycroft et al. (2018).
Abstract (emphasis mine):
Genetic differences within or between human populations (population structure) has been studied using a variety of approaches over many years. Recently there has been an increasing focus on studying genetic differentiation at fine geographic scales, such as within countries. Identifying such structure allows the study of recent population history, and identifies the potential for confounding in association studies, particularly when testing rare, often recently arisen variants. The Iberian Peninsula is linguistically diverse, has a complex demographic history, and is unique among European regions in having a centuries-long period of Muslim rule. Previous genetic studies of Spain have examined either a small fraction of the genome or only a few Spanish regions. Thus, the overall pattern of fine-scale population structure within Spain remains uncharacterised. Here we analyse genome-wide genotyping array data for 1,413 Spanish individuals sampled from all regions of Spain. We identify extensive fine-scale structure, down to unprecedented scales, smaller than 10 Km in some places. We observe a major axis of genetic differentiation that runs from east to west of the peninsula. In contrast, we observe remarkable genetic similarity in the north-south direction, and evidence of historical north-south population movement. Finally, without making particular prior assumptions about source populations, we show that modern Spanish people have regionally varying fractions of ancestry from a group most similar to modern north Moroccans. The north African ancestry results from an admixture event, which we date to 860 – 1120 CE, corresponding to the early half of Muslim rule. Our results indicate that it is possible to discern clear genetic impacts of the Muslim conquest and population movements associated with the subsequent Reconquista.
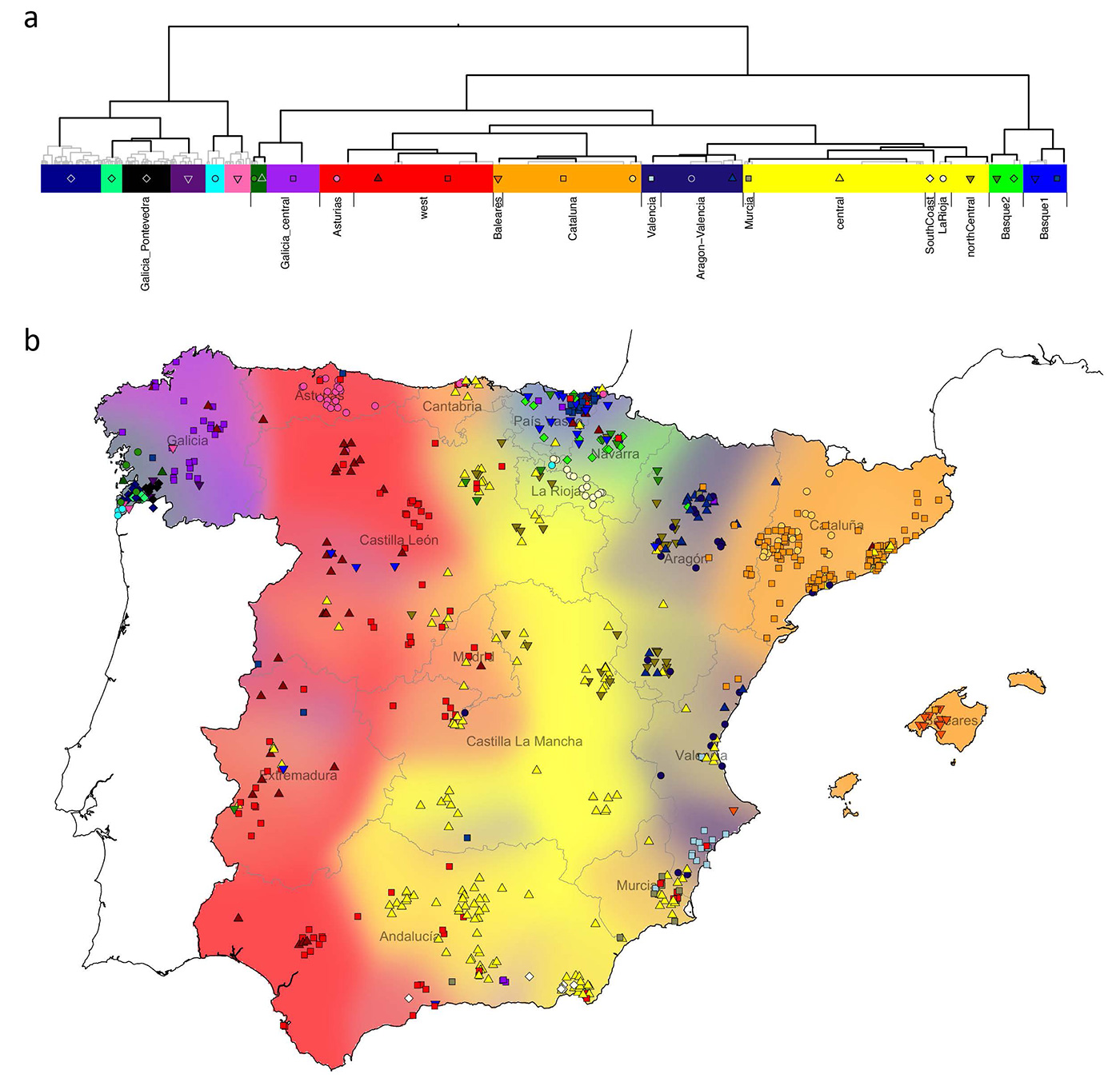
Some interesting excerpts:
Our results further imply that north west African-like DNA predominated in the migration. Moreover, admixture mainly, and perhaps almost exclusively, occurred within the earlier half of the period of Muslim rule. Within Spain, north African ancestry occurs in all groups, although levels are low in the Basque region and in a region corresponding closely to the 14th-century ‘Crown of Aragon’. Therefore, although genetically distinct this implies that the Basques have not been completely isolated from the rest of Spain over the past 1300 years.
NOTE. I must add here that the Expulsion of Moriscos is known to have been quite successful in the old Crown of Aragon – deeply affecting its economy – , in contrast with other territories of the Crown of Castille, where they either formed less sizeable communities, or were dispersed and eventually Christened and integrated with local communities. For example, thousands of Moriscos from Granada were dispersed following the War of Alpujarras (1567–1571) into different regions of the Crown of Castille, and many could not be later expelled due to the locals’ resistance to follow the expulsion edict.
Perhaps surprisingly, north African ancestry does not reflect proximity to north Africa, or even regions under more extended Muslim control. The highest amounts of north African ancestry found within Iberia are in the west (11%) including in Galicia, despite the fact that the region of Galicia as it is defined today (north of the Miño river), was never under Muslim rule and Berber settlements north of the Douro river were abandoned by. This observation is consistent with previous work using Y-chromosome data. We speculate that the pattern we see is driven by later internal migratory flows, such as between Portugal and Galicia, and this would also explain why Galicia and Portugal show indistinguishable ancestry sharing with non-Spanish groups more generally. Alternatively, it might be that these patterns reflect regional differences in patterns of settlement and integration with local peoples of north African immigrants themselves, or varying extents of the large-scale expulsion of Muslim people, which occurred post-Reconquista and especially in towns and cities.
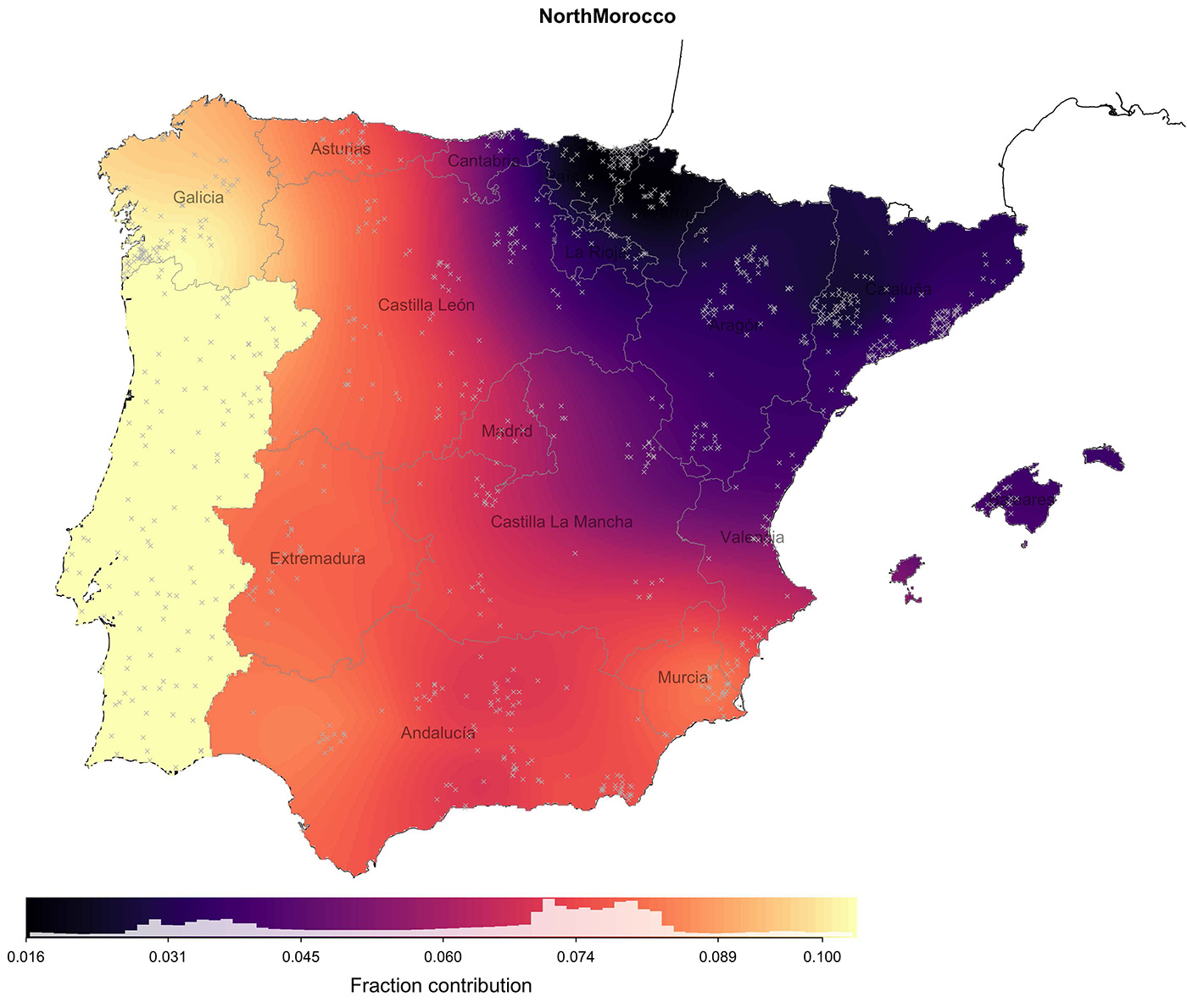
the locations of sampled individuals used in the estimation. Map shows the fraction contributed from the donor group ‘NorthMorocco’.
Overall, the pattern of genetic differentiation we observe in Spain reflects the linguistic and geopolitical boundaries present around the end of the time of Muslim rule in Spain, suggesting this period has had a significant and long-term impact on the genetic structure observed in modern Spain, over 500 years later. In the case of the UK, similar geopolitical correspondence was seen, but to a different period in the past (around 600 CE). Noticeably, in these two cases, country-specific historical events rather than geographic barriers seem to drive overall patterns of population structure. The observation that fine-scale structure evolves at different rates in different places could be explained if observed patterns tend to reflect those at the ends of periods of significant past upheaval, such as the end of Muslim rule in Spain, and the end of the Anglo-Saxon and Danish Viking invasions in the UK.
Certain people want to believe (well into the 21st century) into ideal ancestral populations and ancient ethnolinguistic identifications linked to one’s own – or the own country’s dominant – ancestral components and Y-DNA haplogroup.
We are nevertheless seeing how mainly the most recent relevant geopolitical events and late internal migratory flows have shaped the genetic structure (including Y-DNA haplogroup composition) of modern regions and countries regardless of its population’s actual language or ethnic identification, whether (pre)historical or modern.
Another surprise for many, I guess.
Related:
- Analysis of R1b-DF27 haplogroups in modern populations adds new information that contrasts with ‘steppe admixture’ results
- Migrations painted by Irish and Scottish genetic clusters, and their relationship with British and European ones
- Y-DNA relevant in the postgenomic era, mtDNA study of Iron Age Italic population, and reconstructing the genetic history of Italians
- The Tollense Valley battlefield: the North European ‘Trojan war’ that hints to western Balto-Slavic origins
- Olalde et al. and Mathieson et al. (Nature 2018): R1b-L23 dominates Bell Beaker and Yamna, R1a-M417 resurges in East-Central Europe during the Bronze Age