Josif Lazaridis tweets about an interesting preprint at BioRxiv (eclipsed by today’s Nature papers), Ancestral heterogeneity of ancient Eurasians, by Daniel Shriner.
Abstract:
Supervised clustering or projection analysis is a staple technique in population genetic analysis. The utility of this technique depends critically on the reference panel. The most commonly used reference panel in the analysis of ancient DNA to date is based on the Human Origins array. We previously described a larger reference panel that captures more ancestries on the global level. Here, I reanalyzed DNA data from 279 ancient Eurasians using our reference panel, finding substantially more ancestral heterogeneity than has been reported. This reanalysis provides evidence against a resurgence of Western hunter-gatherer ancestry in the Middle to Late Neolithic and evidence for a common ancestor of farmers characterized by Western Asian ancestry, a transition of the spread of agriculture from demic to cultural diffusion, at least two migrations between the Pontic-Caspian steppes and Bronze Age Europe, and a sub-Saharan African component in Natufians that localizes to present-day southern Ethiopia.
Excerpt (emphasis mine)
Early to Middle Bronze Age Steppe Peoples
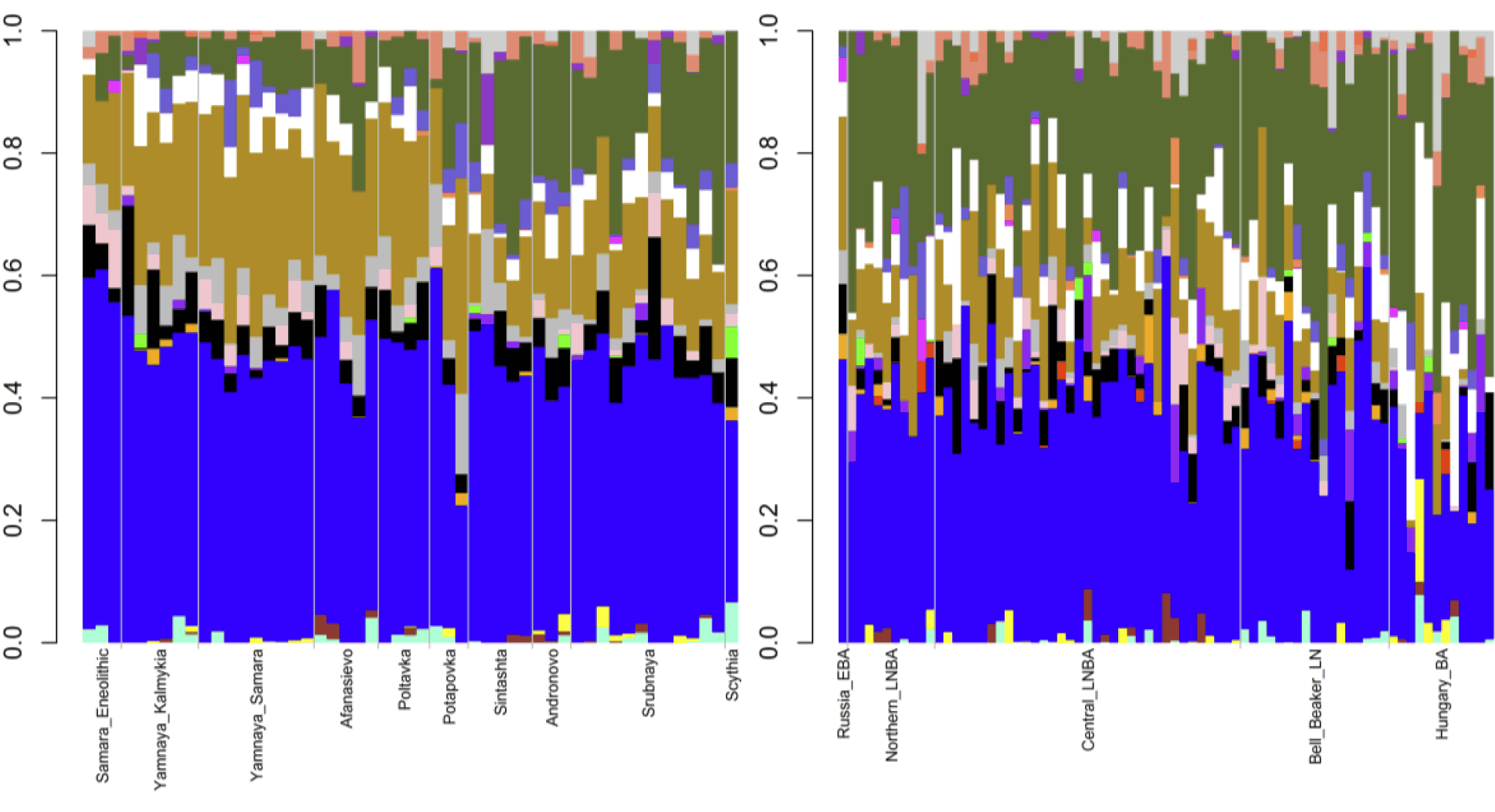
Third, we considered the Eurasian steppe peoples (See figure). The Eneolithic Samara sample had 64.4% Northern European, 18.2% Southern Asian, 8.8% Circumpolar, 4.3% Amerindian, and 4.3% Southern European ancestries. The 27 Early to Middle Bronze Age steppe individuals (Yamnaya from Kalmykia, Yamnaya from Samara, Afanasievo, Poltavka, and Potapovka) averaged 54.7% Northern European, 27.8% Southern Asian, 7.9% Southern European, 4.7% Kalash, 4.2% Amerindian, and 0.8% Western Asian ancestries. We included the Potapovka sample here because the sum of absolute differences in ancestry was greater post-Potapovka rather than post-Poltavka. The increases in Southern Asian and Southern European ancestries do not fit with a European hunter-gatherer source and more broadly do not fit with any of the samples, suggesting an unknown source population. Currently, Southern Asian ancestry co-localizes with Y DNA haplogroup L and correlates with Indo-Iranian languages.Although there are no L haplogroups in any of these Early to Middle Bronze Age steppe individuals, the correlation with Indo-Iranian languages strengthens the connection between Early to Middle Bronze Age steppe peoples and the introduction of Indo-European languages into Europe. In the Early to Middle Bronze Age steppe peoples, 83.3% of Y DNA haplogroups were R1b and 85.2% of mitochondrial haplogroups were H, J, T, or U. Thus, Northern European ancestry was primarily associated with R1b in these peoples, rather than with I2 as in the European hunter-gatherers, while the mitochondrial lineages were more diverse than in the European hunter-gatherers but less diverse than in the Early Neolithic peoples.
It is an interesting new approach, in that it takes into account more than just adxmiture components and PCA to assess ancestral populations.
As simplistic and wrong some conclusions may seem from your point of view, you have to take into account what Iain Mathieson had to (sadly) expressly state recently:
ADMIXTURE clusters are not ancestral populations
— Iain Mathieson (@mathiesoniain) February 21, 2018
Related:
- Olalde et al. and Mathieson et al. (Nature 2018): R1b-L23 dominates Bell Beaker and Yamna, R1a-M417 resurges in East-Central Europe during the Bronze Age
- The Indo-European demic diffusion model, and the “R1b – Indo-European” association
- The concept of “Outlier” in Human Ancestry (III): Late Neolithic samples from the Baltic region and origins of the Corded Ware culture
- Science and Archaeology (Humanities): collaboration or confrontation?
- Massive Migrations? The Impact of Recent aDNA Studies on our View of Third Millennium Europe
- Expansion of peoples associated with spread of haplogroups: Mongols and C3*-F3918, Arabs and E-M183 (M81)